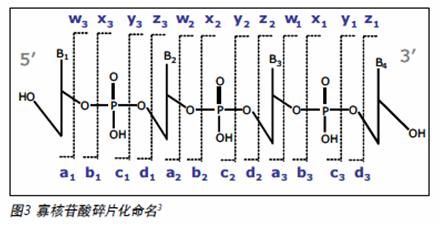
UPLC SYNAPT secondary mass spectrometry method for siRNA oligonucleotide structure confirmation Vera B. Ivleva, Ying Qing Yu and Martin Gilar Milford Waters Corporation, Massachusetts, USA Introduction The RNA interference (RNAi) mechanism plays a fundamental role in post-transcriptional gene silencing. In the case of known sequences, routine silencing of gene silencing can be performed using synthetic RNAi probes of ~21 nucleotides (nt) in length. This method is also used to develop drugs. Synthetic RNAi oligonucleotides must be purified to prevent gene silencing outside the target. RNAi drugs require good experimental descriptions to meet regulatory requirements and minimize the potential for safety and effectiveness. The high resolution chromatographic capabilities and mass spectrometry analysis of Waters Ultra High Performance Liquid Chromatography ( UPLC ® ) technology are powerful tools for the analysis of biopharmaceuticals such as siRNA oligonucleotides. As part of the oligonucleotide confirmation, sequencing can be performed by selective enzymes or chemicals. The secondary mass spectrometry fragmentation method is a rapid general method that can be used for modified oligonucleotides (usually resistant to enzymatic cleavage). In order to obtain structural information on the complete 21-nucleotide RNAi, the molecules in the secondary mass spectrometry must be efficiently ionized and produce sequence-related ions. The mass accuracy and resolution are also appropriate. This application note provides a procedure for accurate UPLC/MS/MS analysis methods for 21-nucleotide RNA structural confirmation. This method is very useful for confirming the sequence of siRNA base drugs. Experimental condition Sample Preparation Complementary RNA strand of 21 nucleotides in length (upstream sequence 5' -UCG UCA AGC GAU UAC AAG GTT -3' , downstream sequence 5' -CCU UGU AAU CGC UUG ACG ATT -3' ) in 0.1 M triethylamine acetate ( Reorganized separately in TEAA). The distribution of synthetic by-products was detected by UPLC/MS together with the upstream and downstream sequences. The sample concentration for UPLC/MS analysis was 30 pmol/μL. method UPLC/MS analysis is consistent with the above. 1 The Waters ACQUITY UPLC ® System is fitted with the ACQUITY UPLC Oligonucleotide Separation Technology ( OST ) C 18 column ( 1.7 μm , 2.1 x 50 mm ; PN 186003949 ). The MS parameters are selected to achieve optimal TEA addition. The deagglomeration effect of the object without affecting the lead ion strength. Waters SYNAPTTM HDMSTM Mass Spectrometry operates in time-of-flight (TOF) mode. The induced ion dissociation collision energy gradient of the selected ions is 25 -55 V. The MS/MS fragmentation is adjusted by selecting the appropriate charge state (-3 to -6) and different collision energy gradients. The product ion spectrum was acquired in the range of 500 - 7000 m / z with a sampling rate of 1 scan / sec. The external calibration of the negative ion mode uses cesium iodide. The map deconvolution before data analysis was performed using MaxEntTM3 software. LC condition LC System: Waters ACQUITY UPLC System Columns: ACQUITY UPLC OST C 18 1.7 μm , 2.1 x 50 mm MS condition MS System: Waters SYNAPT HDMS System Capillary Voltage: 2.7 V Results and discussion The collision energy mode affects the extent to which MS/MS debris is generated and the energy value must be adjusted for the corresponding analyte. In general, the most widely charged states for MS/MS fragmentation for 21 nucleotide length analytes are -5 and -3 (range 1500 - 2000 m /z). A collision energy gradient of 25-45 V is suitable for the generation of fragments available for the 21 nucleotide RNA. In general, to obtain a reasonable signal-to-noise ratio (S/N) for MS/MS fragments, the main component or contaminant in the total ion chromatogram (TIC) must have a signal with a minimum of 500 ions. The complementary RNA21 nucleotide strands were injected into the UPLC/MS/MS system, respectively, and the chromatograms showed shorter oligonucleotide peaks (5' hydrolysates) which were well resolved according to the original target 21 nucleotides ( figure 1 ). The assignment of component peaks is based on their accurate mass measurements and confirms partial sequences. 1 To identify the entire nucleotide structure, the 21-nucleotide downstream strand of the leader ion [M-4H] 4- was isolated and fragmented (Figure 2). The main feature fragments are complementary c and y ions and low intensity [aB] and w ions (named in Figure 3). The ions of the above 5'-PO cleavage sequence are the major fragments of the RNA oligonucleotide MS/MS analysis, which are different from the products produced by DNA (mainly 3'-CO cleavage [aB] and w ions) . This is because DNA molecules lack 2'-hydroxyl groups. 2 The internal fragment produced by the double-stranded framework fragmentation and the cytosine neutral deletion of c- ion (Table 1) were significantly less and did not contribute to the peak assignment of the structure identification. The MS/MS analysis of the upstream 21 nucleotide RNA, the collision energy gradient (30-50 V) yielded similar results (Figure 2). Figure 2 MaxEnt3 deconvolution MS/MS map of a nucleotide RNA oligonucleotide. The asterisk is the harmonic peak (the result of deconvolution) Of the fragment ions in the [M-4H] 4- map of MS/MS, 21 nucleotides of RNA are represented by several charge states (Table 1). The multi-charged ion deconvolution of the MS/MS map is performed by MaxEnt 3 software, which significantly reduces the complexity of the map and simplifies the interpretation of MS/MS data. The deconvoluted map is sufficient to interpret the entire siRNA sequence (Fig. 2), and several peaks representing the loss of A, G and C gas phase nucleobases representing 21 nucleotide molecules are also detected. LockMass has a mass accuracy of less than 10 ppm after calibration and is useful for manual data analysis (Table 1). The mass resolution of the triple quadrupole mass spectrometer and the ion trap instrument may not be sufficient to characterize fragmentation to distinguish multiple charge peaks from other oligonucleotide complex fragment products. in conclusion A method for reliable sequencing by accurate mass UPLC/MS/MS for the confirmation of siRNA oligonucleotide structure was developed. • The high quality accuracy and resolution of the SYNAPT HDMS system achieves clear fragment ion assignment requirements. This highly validated sequencing method is useful for determining pharmaceutically acceptable oligonucleotide sequences according to the US FDA biopharmaceutical compound regulations. This method may also be used for computer to determine unknown impurity sequences. It is expected that this MS/MS method can be used for chemically modified oligonucleotides (step sequencing) where exonuclease is difficult to cleave. Automated MS/MS reduces analysis time in hours or even days, reducing sample analysis costs. In addition, UPLC can quickly isolate target RNA populations, and multiple oligonucleotide strands can perform both MS and MS/MS analysis. The ability of UPLC to isolate a target oligonucleotide from its cleavage product is highly advantageous for siRNA metabolism studies. references 1. UPLC/MS Analysts of Interfering RNA Oligonucleotides. Waterrs Application Note.2008 : 720002412en.
Henan Qianli Machinery CO. Ltd is a profession manufacture of Farm Tractor, rice machine, Reaper Machine, Power Tiller, garden machine and other farm implements. Our factory was established in 1987 and has more than 30 years of production and sales experience. We focus on providing customers with high-quality products and excellent services.
Gas Engine Generator,Gasoline Generator Price,Portable Gasoline Generator,Gasoline Powered Generators Henan Qianli Machinery Co.,Ltd. , https://www.chalionagri.com
Column temperature: 60 °C
Injection volume: 5 μL
Flow rate: 0.2 mL/min
Mobile phase A: 15 mM TEA, 400 mM HFIP
Mobile phase B: 50% A, 50% methanol gradient: B liquid content from 20-40% in 10 minutes
Sample cone voltage: 31 V
Extraction cone voltage: 3 V
Source temperature: 120 °C
Desolvation gas temperature: 300 °C
Desolvent flow rate: 500 L / h
Collision trap energy: 6 V
Transport collision energy: 4 V
Mass resolution: ~ 9000 (FWHH) in V mode
LockMass : CsI 10 mg/mL (water-isopropanol, 1 : 1 ), flow rate 5 μL/min, scan time 1 second, frequency 30 seconds, set mass 1685.765 m / z ( Cs 6 I 7 - ) 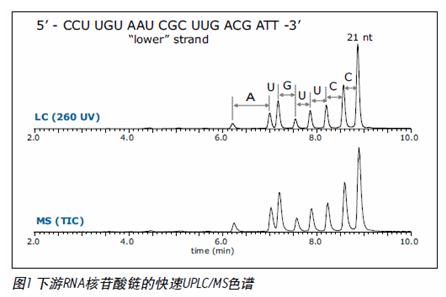

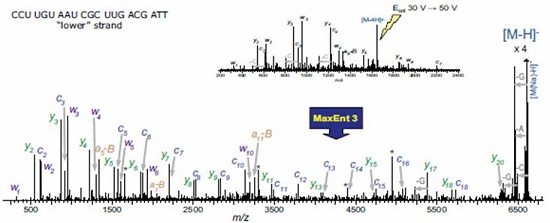
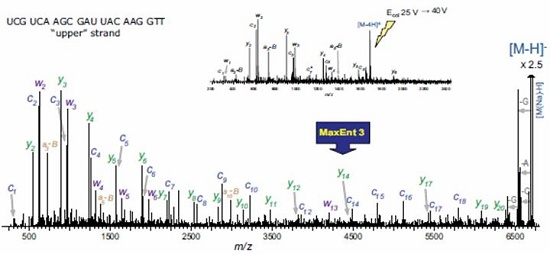
• A reliable MS/MS method produces characteristic ion fragments that cover a wide range of masses and are sufficient for the resolution of 21 nucleotide siRNA sequences.
• MaxEnt 3 deconvolution software greatly reduces the complexity of the MS/MS map and simplifies the sequence resolution process.
2. Kirperkar F, Krogh TN. Rapid Commun. Mass Spectrom. 2001 ; 15 ( 1 ): 8-14.
3. Adopted from McLuckey SA, et al. J. Am. Soc. Mass Spectrom. 1992 ; 3 ( 1 ) 60-70.
UPLC SYNAPT Secondary Mass Spectrometry for siRNA Oligonucleotide Structure Confirmation